It is possible to sequence using the so-called NGS method, which allows a complete reading of the entire virus genome. Thanks to sophisticated software, it is possible to analyze where the virus by which a particular patient is infected comes from and also to detect possible mutations in the virus (so-called lines), i.e. changes in individual parts of the genome.
“If a thorough quality control of the samples is not performed, the subsequently analyzed data may be inaccurate and may form artificial lines of non-existent mutations. Therefore it is very important to monitor the whole process in order to be able to describe the real situation of the spread of the virus in society,” points out one of the creators of the COVID cloud application Petr Brož, CEO of Bioxsys company, which mainly develops software for genetic analysis.
The analysis may reveal new mutations in the virus, their representation and spread in the population
Thanks to the connection of bioinformatics experts from Bioxsys and application laboratory specialists from BioVendor, they were able to quickly offer even to beginning and inexperienced laboratories a complete solution for next generation virus sequencing.
“Together with uniquely designed chemistry, we have created software for the analysis of data from sequencers - devices that allow laboratories to sequence, i.e. read the complete genome of the virus and thus obtain valuable information about changes in the virus and its spread. Enriching primary data with epidemiological information and saving it on a long-term basis is key to creating statistical analyzes that will help fight the epidemic,” explains Brož
The software then, if epidemiological data are consistently filled in, allows the use of statistical methods to detect the possible development of Covid-19 in patients from which the mutation spreads and its ability to escape the immunity of vaccinated patients.
This online software analyzes all necessary data in the context of data from sequencing around the world.
“In the Czech Republic, in addition to well-established institutions such as the Academy of Sciences, CEITEC, the State Institute of Public Health and the University Hospitals, an increasing number of diagnostic laboratories and scientific workplaces are currently engaged in sequencing. Therefore, it is necessary to standardize the quality of the data so that it can be saved in global databases that monitor SARS-CoV-2 mutations,” points out Petr Brož, bioinformatic scientist.
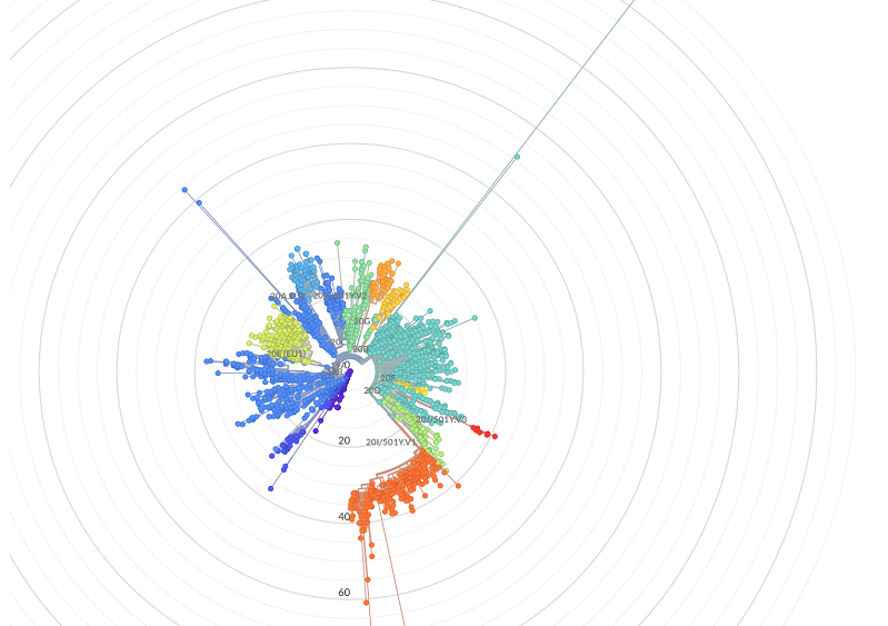
Variants or mutations of the virus are compared with the so-called reference genome from Wuhan (Wuhan-Hu-1). The software can thus compare how much the virus changes over time and where the individual mutations spread, how the Czech Republic is doing in comparison with other countries and how the mutations manifest themselves in society. It creates so-called phylogenetic trees, thanks to which experts can exactly describe individual variants.
Thanks to sequencing, epidemiologists have comprehensive information
“Mutations have been spreading a lot in the population since the autumn, because the virus mutates relatively easily. However, only some of them have a more significant effect on society, for example due to their infectivity or, for example, the fact that infection with this variant is more difficult in most patients,” explains Kateřina Pehlíková, who deals with molecular biology and NGS at the BioVendor Group. Together with the BioVendor team, she is currently helping laboratories with the introduction of the NGS testing method. “Unlike PCR tests, which recognize only a small part of the genome, the NGS method is a complete reading of the entire genome, so we have much more information about it,” she explains.
Thanks to an online tool developed by both companies, epidemiologists can track the exact spread of individual mutations in society, their origin, and the rate of transmission and response of patients. “However, this requires a sufficient amount of data from testing. At BioVendor, we are currently helping laboratories to implement this new method, thanks to the setting of criteria for quality control and new software that works with data directly from the sequencer, sufficient data quality is guaranteed,” points out Pehlíková from BioVendor.
The tool then allows to share the data to global databases, making it possible to compare and analyze local data in a global context. “If we are able to identify that, for example, all the dead have the same virus mutation, we can develop a simple PCR test, a test kit, directly for this variant and detect it in time. PCR tests can detect the presence of SARS-CoV-2 virus, or search for a specific variant, but we must know the exact information about each mutation,” explains the use of analyzes Pehlíková from BioVendor.
The aim of the application is to simplify and automate the analysis of data from sequencing for laboratories as much as possible, while also ensuring that low-quality data does not enter the databases and thus does not distort the statistics of the occurrence and spread of individual virus mutations.




